Research in BRAINSTRUC
The overarching goal of BRAINSTRUC is to understand the molecular basis of selected brain functions and diseases through spatiotemporal structural models of complex biomolecular systems.
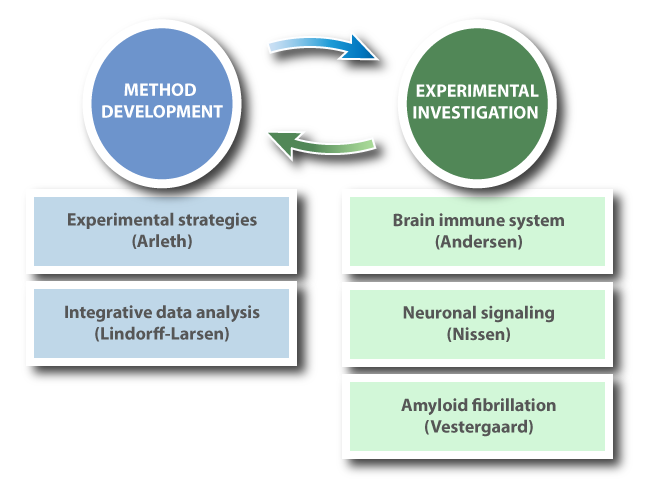
The BRAINSTRUC consortium is focusing on two overall goals:
- To develop improved methods for integrated structural biology data collection and data interpretation. This development will be driven by highly complex questions from the three neurobiological systems.
- To investigate the structural aspects of selected systems that are central to neuronal signalling, inflammation and disease. This investigation will be enabled by the significant methodological development.
Goal 1: Theoretical and Methodological Developments
The first goal is necessitated by the complexity and size of the biological systems studied in BRAINSTRUC. It will include further developments of data-integration and simulation methods as well as experimental developments. A particular focus is on the exploitation of the novel opportunities offered at the new large-scale infrastructures at the European Spallation Source (ESS) and MAX-IV in Lund, as well as the recent revolutionary development within cryo-electron microscopy.
- Integrative data analysis will be implemented to develop a set of algorithms, encoded in scientific software, which enable new approaches in dynamic integrative structural biology for large and complex systems. This in turn provides us with new opportunities to tackle much more difficult biological problems. Examples include those described below under goal 2, but the approaches will be generally applicable also outside BRAINSTRUC.
- Experimental method development to improve sample quality of membrane proteins for SAXS/SANS, MX/XRD and cryoEM/ET will enable high resolution information to be obtained from such systems. Furthermore, obtaining insight into dynamics is crucial both for understanding function of macromolecules and for decomposing an ensemble of structural states. Methods to generate this type of information will also be developed to fully exploit the emerging opportunities at MAX IV and e-XFEL. Finally, systematic and strategic approaches for integrative data analysis of complex biostructural problems will be developed to support the study of such systems, exemplified by the examples below.
Goal 2: Structual investigations
Results from the first goal, will be used to adress the second goal through the investigation of three central and highly complex biomolecular systems involved in neuronal signalling, inflammation and disease. Structural studies of these systems represent significant experimental challenges:
- The complement system plays highly important roles in brain inflammation, neuronal development, homeostasis and neurological diseases such as neuromyelitis optica (NMO) and myastenia gravis (MG) where an autoimmune response involving complement activation causes severe damage. Complement is part of innate immunity and comprises more than 50 proteins that are also produced locally in the brain. Complement activation progresses through the formation of very large intricate and dynamic multi-component protein complexes greatly complicating their structural analysis.
- Neuronal signaling involves convoluted interactions between multiple membrane transport and channel proteins, regulating ion fluxes, such as the P-type ATPase ion pumps and cation-chloride transporters, and consequently several neurological diseases associate with their function or dysfunction. Similarly, the transport processes maintaining asymmetric lipid distributions potentiate neuronal membranes for their role in membrane-associated and vesicle-mediated signaling. The higher-order structures of neuronal membrane proteins play intriguing roles in signal transduction along axons. The structural basis of neuronal signaling including the structural organization of membrane proteins and (membrane-)protein:protein interplay constitute one of the most challenging questions in structural biology today.
- Protein amyloid fibrillation is intimately associated with the highly prevalent neurodegenerative diseases, such as Parkinson’s (PD) and Alzheimer’s (AD) diseases. The structural basis of the cytotoxic effect is, however, far from understood. Protein fibrillation is inherently difficult to study structurally, due to the multiple, heterogeneous structural events leading to the formation of fibrils, involving a variety of protein structural changes and multiple transient, dynamic protein:protein interactions.
Perspectives
By using the most advanced techniques available in structural biology BRAINSTRUC will provide significant insight into the molecular basis of three selected aspects of neuronal/glial function and dysfunction.
As described, the biological focus will necessitate and drive the method development within BRAINSTRUC. This will result in new tools that are central for obtaining the anticipated biological results. In addition, these novel methods and tools will be highly valuable for the broader structural biology community, thereby stimulating further to novel achievements, also beyond the scope of BRAINSTRUC.
BRAINSTRUC also has an important mission to significantly push the frontiers globally within structural biology and in particular to train young scientists to continue the strong tradition for excellence in Danish structural biology. BRAINSTRUC will allow its members to take maximum advantage of nearby world-class research infrastructure (ESS, MAX IV, eXFEL). A close interplay with staff at these new and developing facilities promises to increase mutual benefits and hence overall value of the large investments in the region.
Funding

BRAINSTRUC was a Lundbeck Foundation initiative on integrative structural biology funded by a 5 year grant.